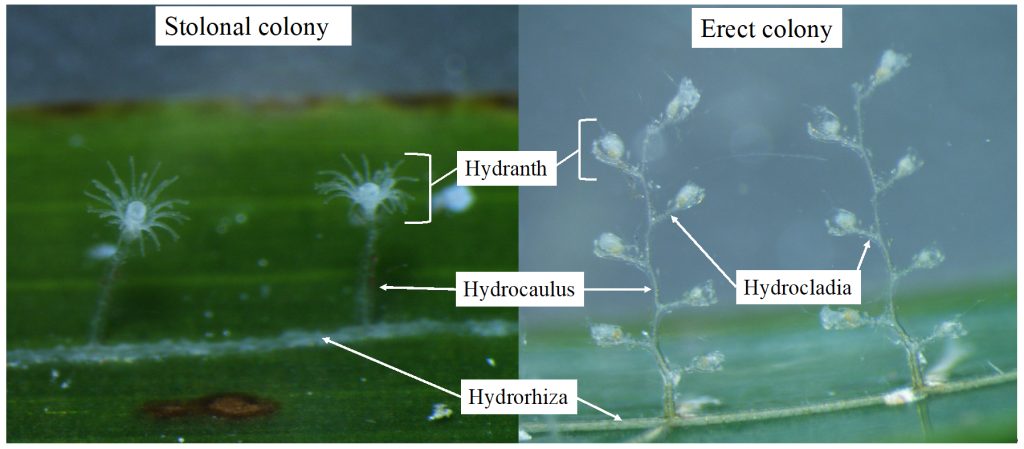
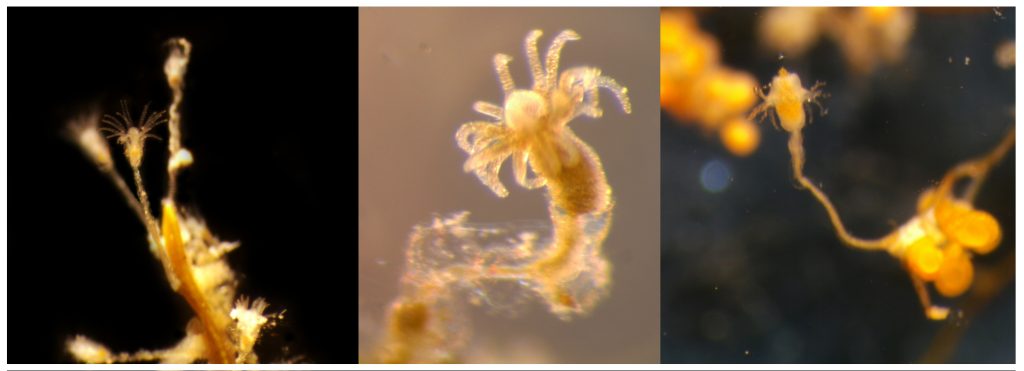
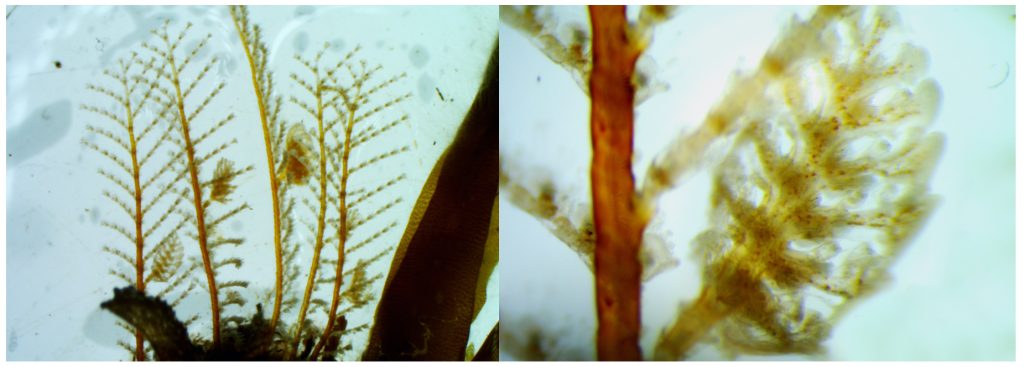
ParaZoo (complete name ‘Metazoan parasites of non-crustacean zooplankton’) is one of the most interaction-focused projects currently running at the Invertebrate Collections of our Museum. This project, funded by the Norwegian Biodiversity Information Centre (Artsdatabanken), aims at studying the different animals that live together inside and on the surface of Norwegian jellyfish. This means that for the next two years we will be looking for tapeworms, flukes, roundworms, and amphipods as ParaZoo tries to answer the question of which of these organisms are associated with gelatinous hosts in Norwegian waters.
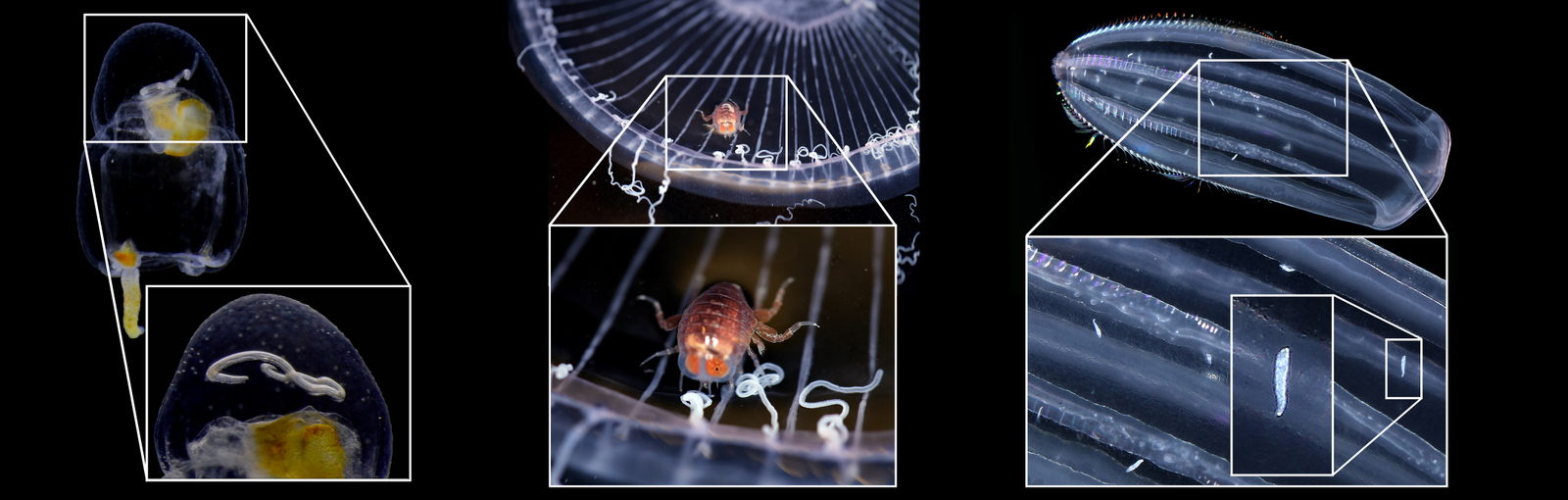
ParaZoo is focused on animal parasites and symbionts associated with jellyfish. Thse parasites include roundworms like Hysterothylacium aduncum (left, in Euphysa aurata), amphipods like Hyperia medusarum (middle, on Aequorera forskalea), and flukes like the members of family Didymozooidae (right, in Beroe gracilis). IC: Aino Hosia (left), Katrine Kongshavn (middle), Joan J. Soto-Àngel (right).

Besides jellyfish, arrow worms (Chaetognatha) are also members of non-crustacean zooplankton that host different types of parasites, like this H. aduncum roundworm (Nematoda). IC: Joan J. Soto-Àngel, Luis Martell.
Parasitism and symbiosis are extremely common life styles in the animal kingdom. In fact, some researchers believe that there may be more species of parasites than of free-living animals, given that each free-living species hosts many species of parasites (most of them unique) and those parasites also host their own parasitic tenants. Marine zooplankton is no exception to this trend, and many parasites and symbionts are expected to occur in copepods, krill, and gelatinous zooplankton. Jellyfish and arrow worms, for example, may be important hosts for flatworms and other helminths, yet our knowledge of these animals in Norway is very scarce.

ParaZoo’s logo includes two of the target taxa of the project: roundworms (Nematoda) and hyperiids (Amphipoda). The third main parasitic group covered is flatworms (Platyhelminthes), illustrated by the larvae of Derogenes varicus parasitizing Halopsis ocellata shown in the right side of this figure. IC: Joan J. Soto-Àngel, Luis Martell.
Understanding zooplankton parasites is important because many of them are going to be transmitted to fish, where they may cause serious diseases. To get a better overview of which critters live in non-crustacean zooplankton, ParaZoo will sample, record, and DNA-barcode specimens from all over the country. The collected animals will be included in our museum collections after being identified, documented photographically, and fixed in ethanol. We will then generate an open-access database of information including pictures and DNA sequences that will help with the identification of the parasites. Aquaculture facilities, fishermen, and managers of marine areas will benefit from this database to better plan and counter potential negative impacts caused by the parasites.

Flukes, such as Opechona spp, parasitize gelatinous zooplankton (in this case a sea-gooseberry Pleurobrachia pileus) as larvae called metacercariae. IC: Joan J. Soto-Àngel, Luis Martell.

The larvae of tapeworms (Cestoda) sometimes use jellyfish to reach their definitive hosts: fish. IC: Joan J. Soto-Àngel, Luis Martell.
ParaZoo is committed to present the diversity of jellyfish parasites to all those not familiar with them. In order to do that, we will regularly write entries here on the blog, as well as participate in several academic and not-academic meetings. The official info webpage for the project is available here, so don’t forget to check it out!
–Luis