It’s funny to see the different reactions to fresh material that comes in to the museum; the exhibition team had received some kelp that will be pressed and dried for the new exhibitions (opening fall 2019), and I ducked in to secure some of the fauna sitting on the kelp before it was scraped off and discarded. For the botanists, the animals were merely a distraction that needed to be removed so that they could deal with the kelp, whilst I was trying to avoid too much algae in the sample as it messes up the fixation of the animals.
- Big buckets of kelp
- Evaluating some of the kelp – how to press it? The stipes (“stem”) presents some challenges, but is seems to work!
- There is a multitude of epiphytes on the stipes, both algae and animals
I then spirited my loot into the lab, and set up camp.
Count me in amongst the people who stare at lumps of seaweed.
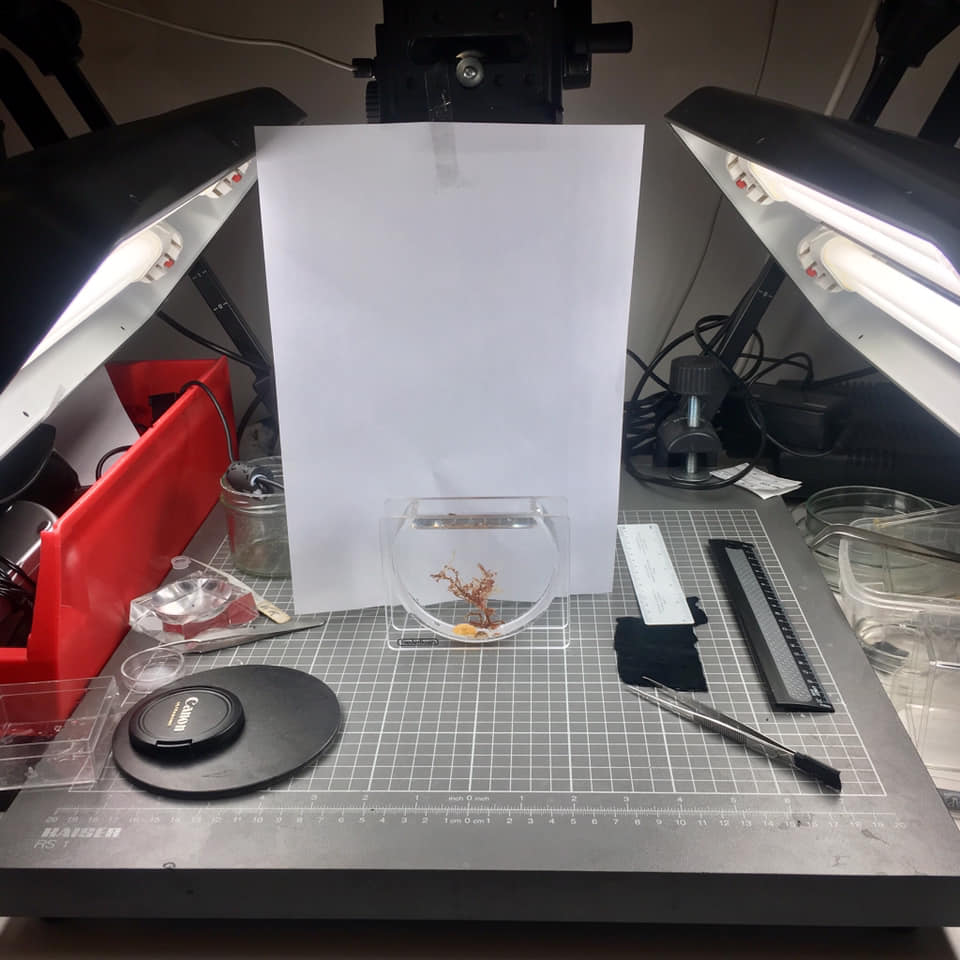
- Photo set-up. Picture by C. Rauch
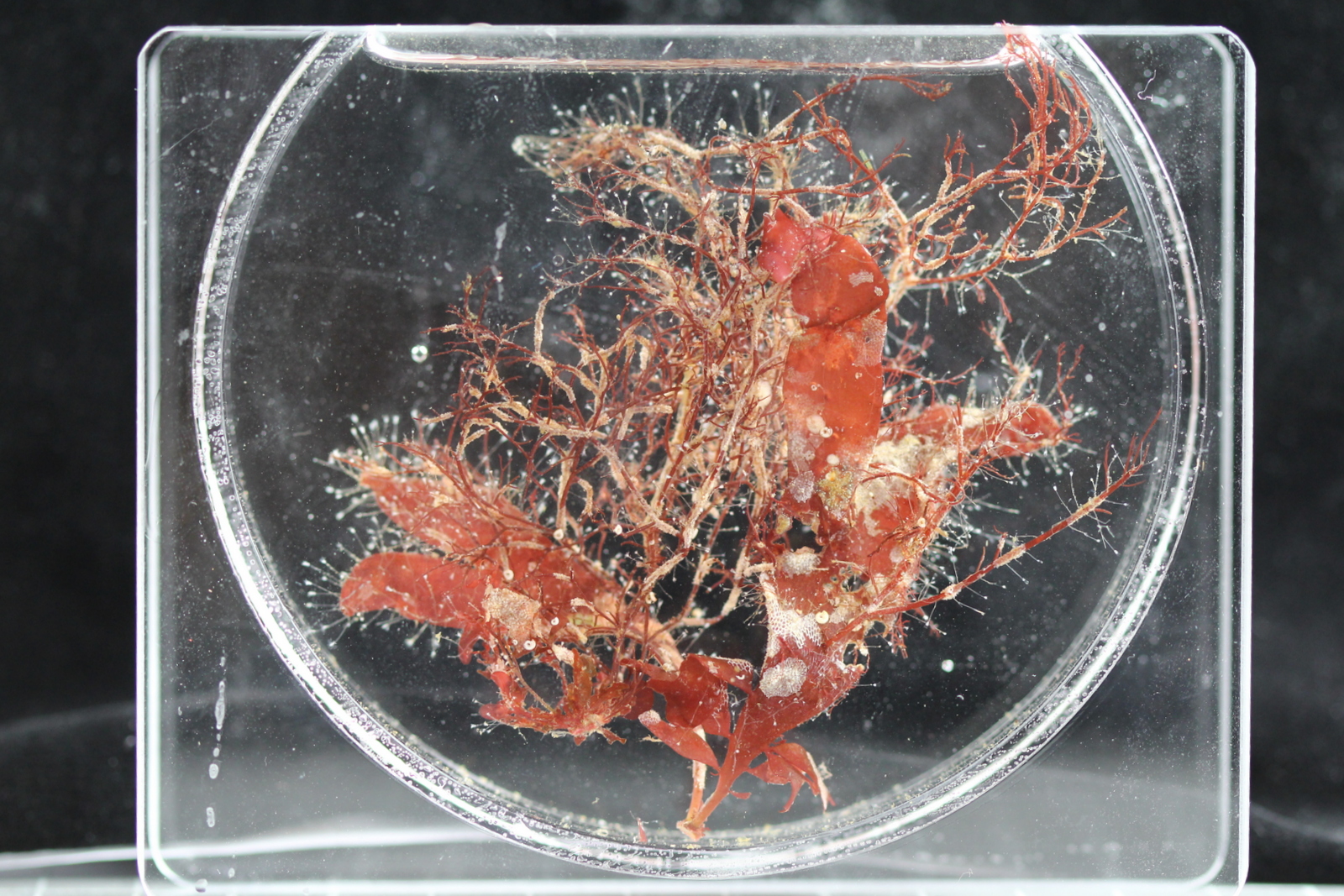
- Nudibranchs and kelp. Picture by C. Rauch
How many animals do you see here? Which ones appeal to you?
I have made a quick annotation of some of the biota here:
Let’s go closer on a small piece of algae:
For Luis, the first thing to catch the eye was (of course) the Hydrozoa

Hydrozoans (the christmas light looking strings), encrusting bryozoans (the flat, encrusting growth on on the algae – you might also know them as moss animals), and some white, spiralling polychaete tubes (photo: K. Kongshavn)
Did you spot the sea hare (Aplysia punctata?) Look a bit above the middle of the photo of the tiny aquarium with the black background. Do you see a red-pink blob?

Hello, Aplysia punctata! (photo: K. Kongshavn)
There were also several other sea slugs that I have handed over to Cessa for inclusion in the sea slugs of Southern Norway project, here are a few:
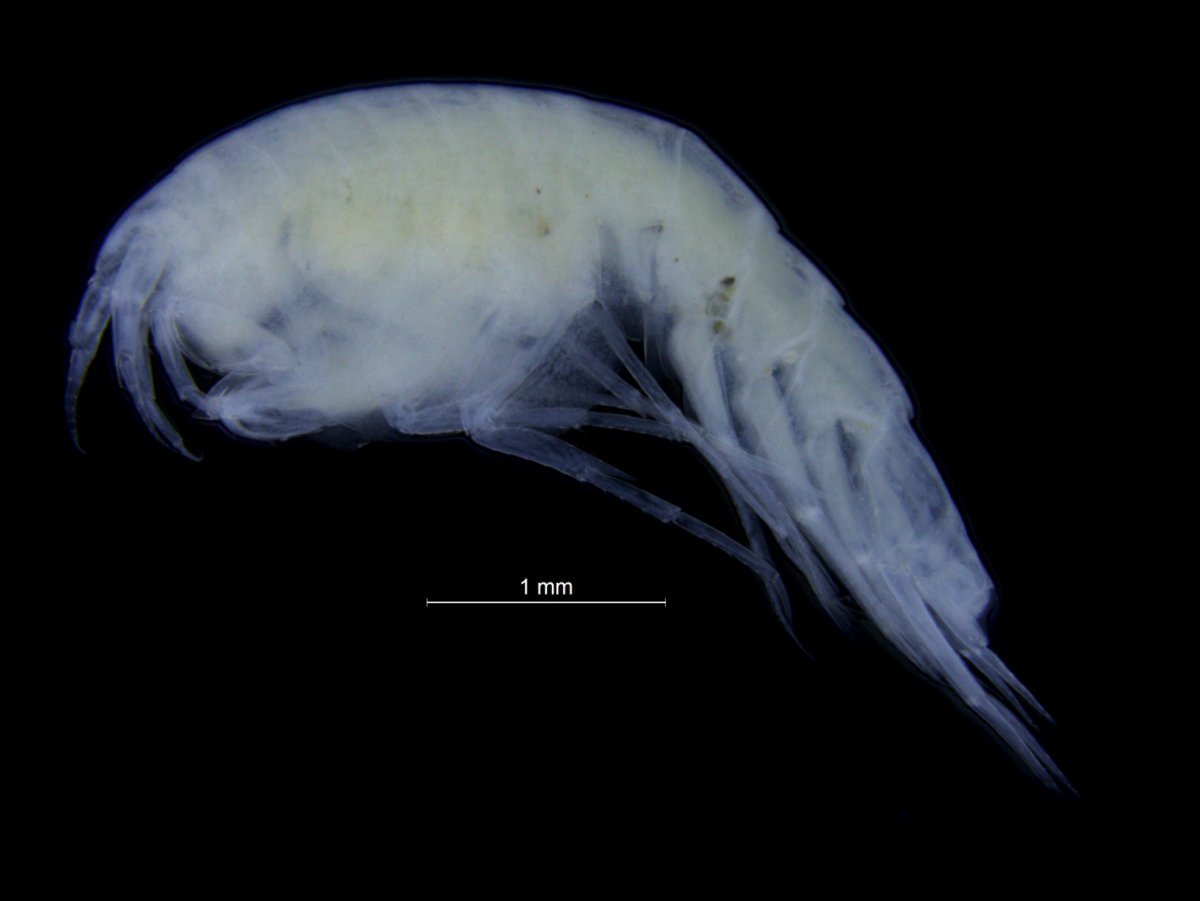
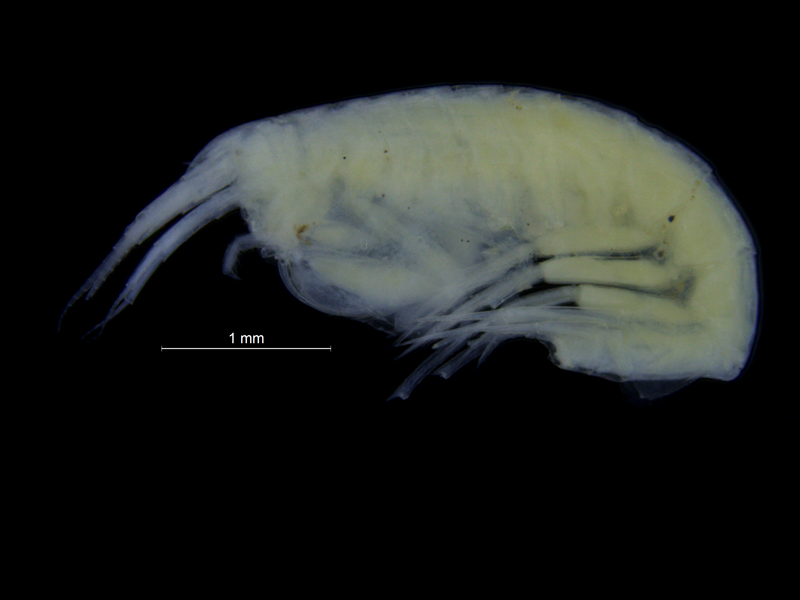
- Tiny nudibranch, about 3 mm long
- Bigger nudi, this one was 2 cm and bright yellow – not too hard to see!
- Catch of the day: a nudibranch that the Sea slugs of Southern Norway project so far only had one record of! It is only 4 mm long, so it took some staring to find it..!
Then there were the shelled gastropods:
- Hi!
- Not sure if this is a different species or just another colouring?
- Tiny, tiny gastropod on bryozoa

The brittle star from the earlier image – this is a Ophiopholis aculeata, the crevice sea star (photo: K. Kongshavn)

In fact, they both are Ophiopholis aculeata (in Norwegian we call them “chameleon brittle stars” – they live up to the name!), one of the very common species around here. (photo: K. Kongshavn)
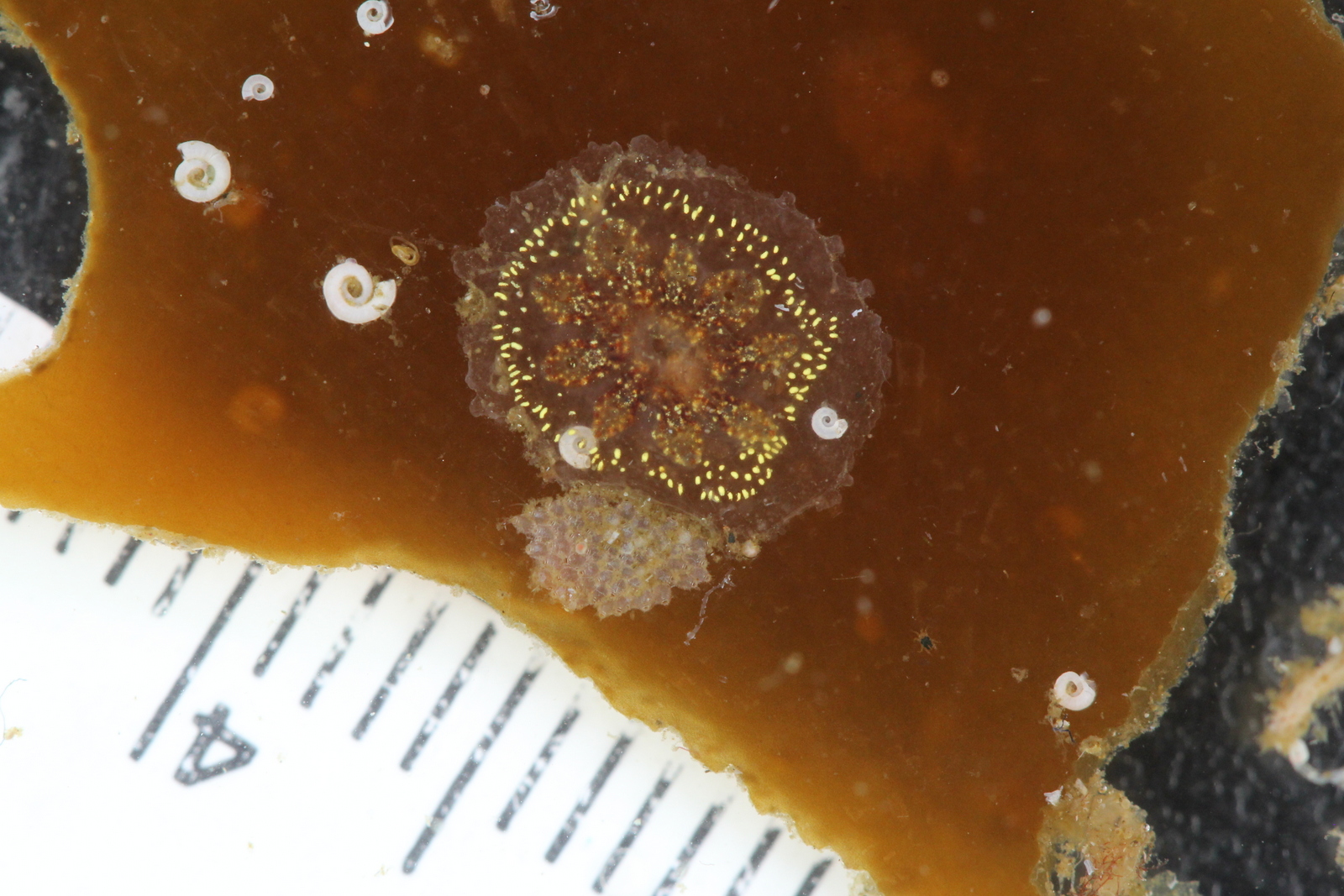
One of the colonial ascidian tunicates (and some of the ever present bryozoa just below it) (photo: K. Kongshavn)
Most of these animals will be barcoded, and will help build our reference library for species that occur in Norway. I also hope that they may have helped open your eyes to some of the more inconspicuous creatures that live just beneath the surface?
2019 will see the start of a new species taxonomy project where we will explore the invertebrate fauna of shallow-water rocky shores, so there will be many more posts like this to come!
-Katrine