In a previous blog post I explained briefly how DNA-sequences are produced for the DNA-barcode library. Now I will show how the BOLD database can be utilized to identify species from sequences.
Say you have access to a lab that can produce DNA-sequences and you have a sample of a crab you cannot identify because some of the key characters are on body parts that have been broken and lost. You produce a DNA-sequence from the “barcode-gene” and open the identification engine in BOLDSYSTEMS.org.
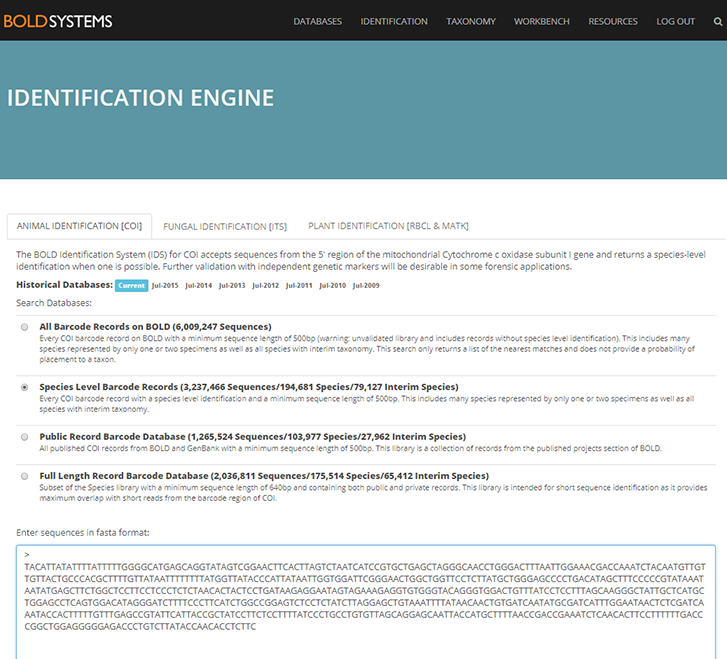
Internet start window for the BOLD identification engine where you paste your unknown DNA sequence into the bottom blank window. (Click on picture to expand)
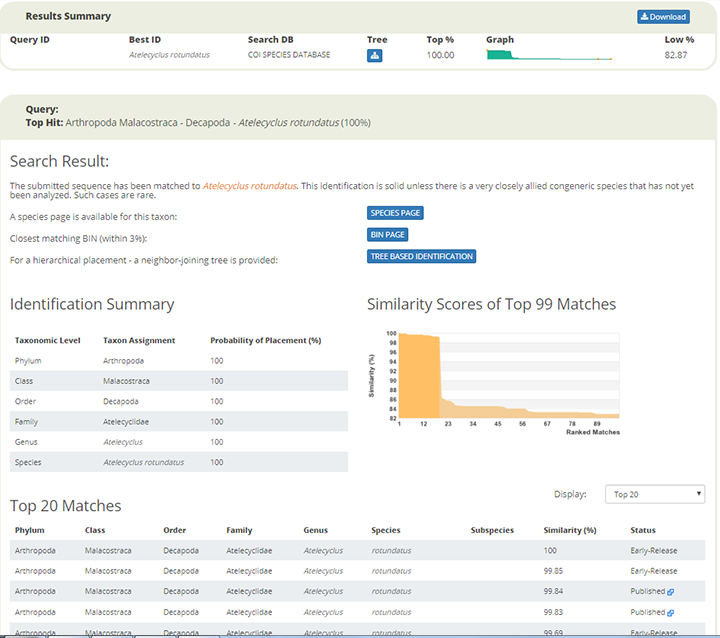
Having submitted your query to BOLD, you wait for some seconds for results. In this example BOLD returned the following window.
The results window lists the top matches in terms of sequence similarity, and in this case we have 100 % similarity match with the crab Atelecyclus rotundatus. There is also an option to display the results as a TREE BASED IDENTIFICATION. When clicking on the option tab, the closest hits are clustered in a so-called Neighbour Joining Tree. In the window below you see parts of the tree where our unknown DNA-sequence has been joined to a group of other sequences in BOLD that have been deposited as Atelecyclus rotundatus barcodes by other biodiversity labs.
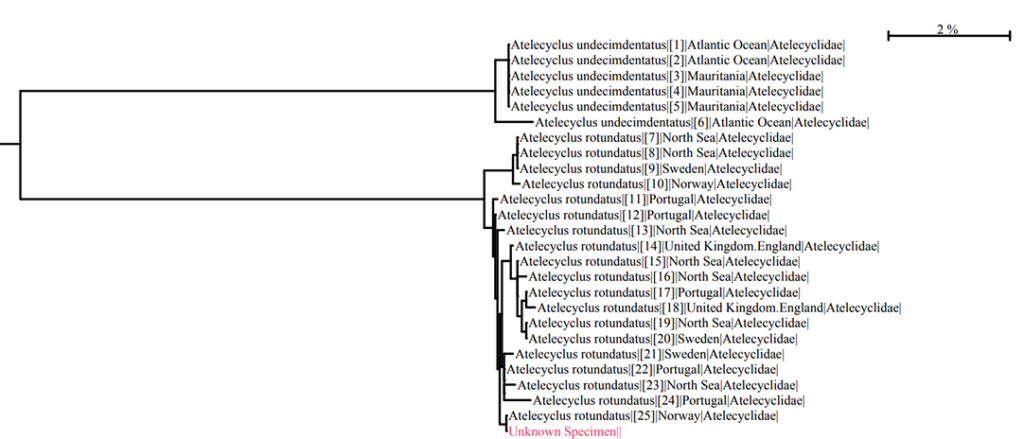
Part of TREE BASED IDENTIFICATION of an unknown DNA sequence (in red). We see that the unknown clusters with with other sequence of Atelecyclus rotundatus. The nearest neighbour branch is Atelecyclus undecimdentatus. (Click on picture to enlarge.)
The species page for Atelecyclus rotundatus gives us more information about this crab and about its records in BOLD.

Species page for the individual we identified with the BOLD identification engine. (Click picture to enlarge.)
If in fact your sequence was produced from an unknown crab, this identification seems convincing. But sometimes you should think twice about search results, and this will be the topic of a future blog post.
-Endre